Visualisation of Blood Group Antigens
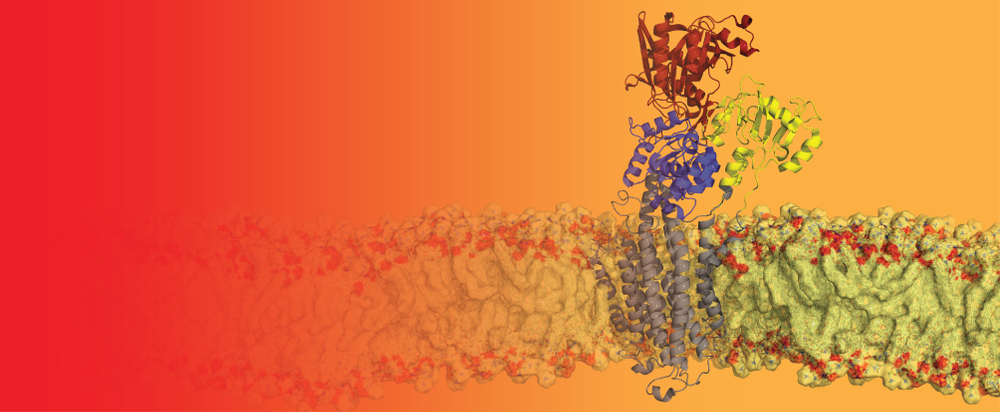
In humans, around 40 different blood groups have been described, a vast excess of the widely known ABO and Rh systems. Hundreds of different genetic (allelic) variants within the genes that define our blood groups have been found, many of these with significant medical implications, due to their potential to cause adverse immune responses of due to links with certain diseases. I am interested in visualising and analysing such variations on the molecular level, by generating models of various important red blood cell surface proteins and mapping the position of key antigens. Molecular dynamics (MD) simulations allow us to explore the behaviour of the proteins in their native membrane environment.
Read more here: https://pubmed.ncbi.nlm.nih.gov/39726599/
Bacterial Lipid Flippases
Bacteria constantly remodel and re-shape their membranes, utinizing a complex machinery of enzymes, in order to survive in an ever-changing environment. One particularly fascinating enzyme, MprF (Multiple peptide resistance Factor), modifies the head group of bacterial membrane lipids and then transports them to the outer leaflet, thereby altering the surface charge of the bacterium. This can give them an advantage when challenged by harsh conditions or certain anti-microbial defense mechanisms or drugs. We have recently characterised MprF from the human pathogen Pseudomonas aeruginosa by determining its structure by cryo-EM and by a thorough functional characterization in lipid vesicles.
Read more here: https://www.science.org/doi/10.1126/sciadv.ads9135
Fungal Proton Pumps
A main focus of my work at the University of Oxford was on primary active transport proteins from the P-type ATPase family. P-type ATPases are highly flexible multi-domain transporters that use energy from ATP hydrolysis for the specific transport of ions across cellular membranes. We are particularly interested in the ATPase with the smallest substrate of all, the H+-ATPase – or proton pump.

It pumps single protons across the plasma membrane in plants and fungi and thereby creates the proton-motive force, which is vital for the organism i.e. by fueling vital nutrient uptake systems.In certain fungi, the proton pump forms hexamers of ~600 kDa.
We study the structural basis for proton transport process and its regulation using a variety of methods and targets.
Due to the fact that the proton pump is essential for all fungi, this research provides an important framework for the development of novel antifungal medication.
Read more about our work on Pma1 here: https://pubmed.ncbi.nlm.nih.gov/37741574/
https://pubmed.ncbi.nlm.nih.gov/34757782/
Methodology
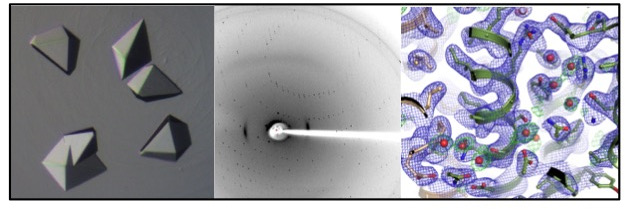
My core expertise lies within protein structure determination methods such as X-ray crystallography, electron microscopy, functional characterization by liposome reconstitution, transport and ATPase assays, and in silico analyses such as MD simulations.
Through successful collaborations our studies also feature nanodiscs, native mass spectrometry and structure-based drug discovery.